Dr. Lubin's research interest is focused on understanding the cellular and molecular mechanisms underlying transcriptional regulation of genes in the nervous system to mediate cognition and how these mechanisms are altered with aging and cognitive disorders including epilepsy and Alzheimer's disease. Currently, our research is directed at characterizing the role of epigenetic mechanisms, such as histone methylation, DNA methylation, non-coding RNAs, and the interaction of the NF-kB signaling pathway with chromatin to determine how these molecular changes participate in the regulation of gene expression related to the normal process of learning and memory and how they are deranged with cognitive deficits. Lubin Review
Dr. Lubin's research interest is focused on understanding the cellular and molecular mechanisms underlying transcriptional regulation of genes in the nervous system to mediate cognition and how these mechanisms are altered with aging and cognitive disorders including epilepsy and Alzheimer's disease. Currently, our research is directed at characterizing the role of epigenetic mechanisms, such as histone methylation, DNA methylation, non-coding RNAs, and the interaction of the NF-kB signaling pathway with chromatin to determine how these molecular changes participate in the regulation of gene expression related to the normal process of learning and memory and how they are deranged with cognitive deficits. Lubin Review
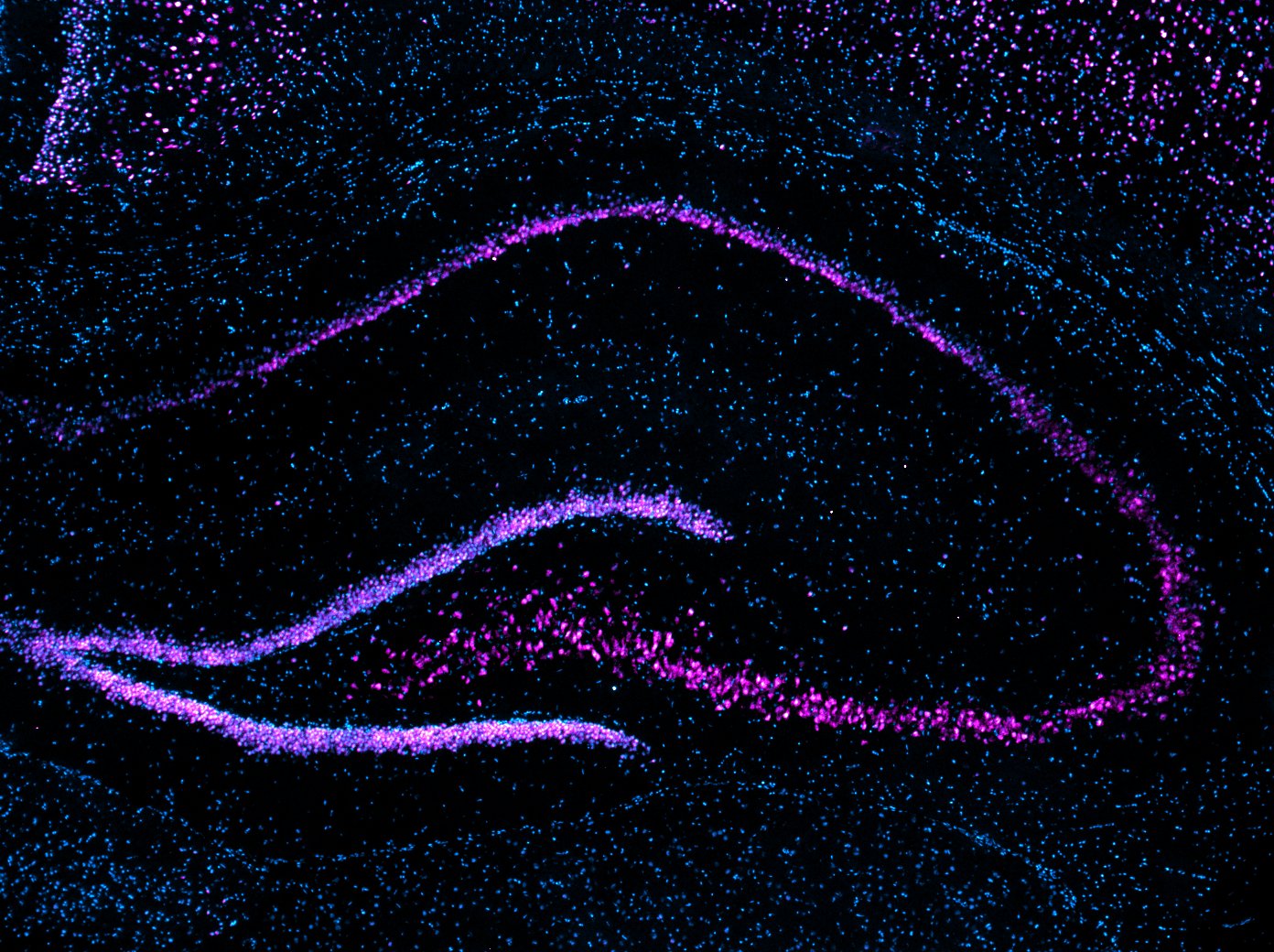
Our research program focuses on neurons and synapses in the hippocampus, entorhinal cortex, and amygdala, brain regions that play an important role in learning and memory and in the development of several cognitive deficits in humans. We have found that memory-associated gene transcripts are dynamically regulated by DNA methylation and specific histone lysine methylation changes in the hippocampus during memory consolidation. Current work includes an assessment of DNA/Histone demethylating agents and viral/chemogenetic tools to manipulate function of the epigenome that may be promising in the mitigation of cognitive disorders.

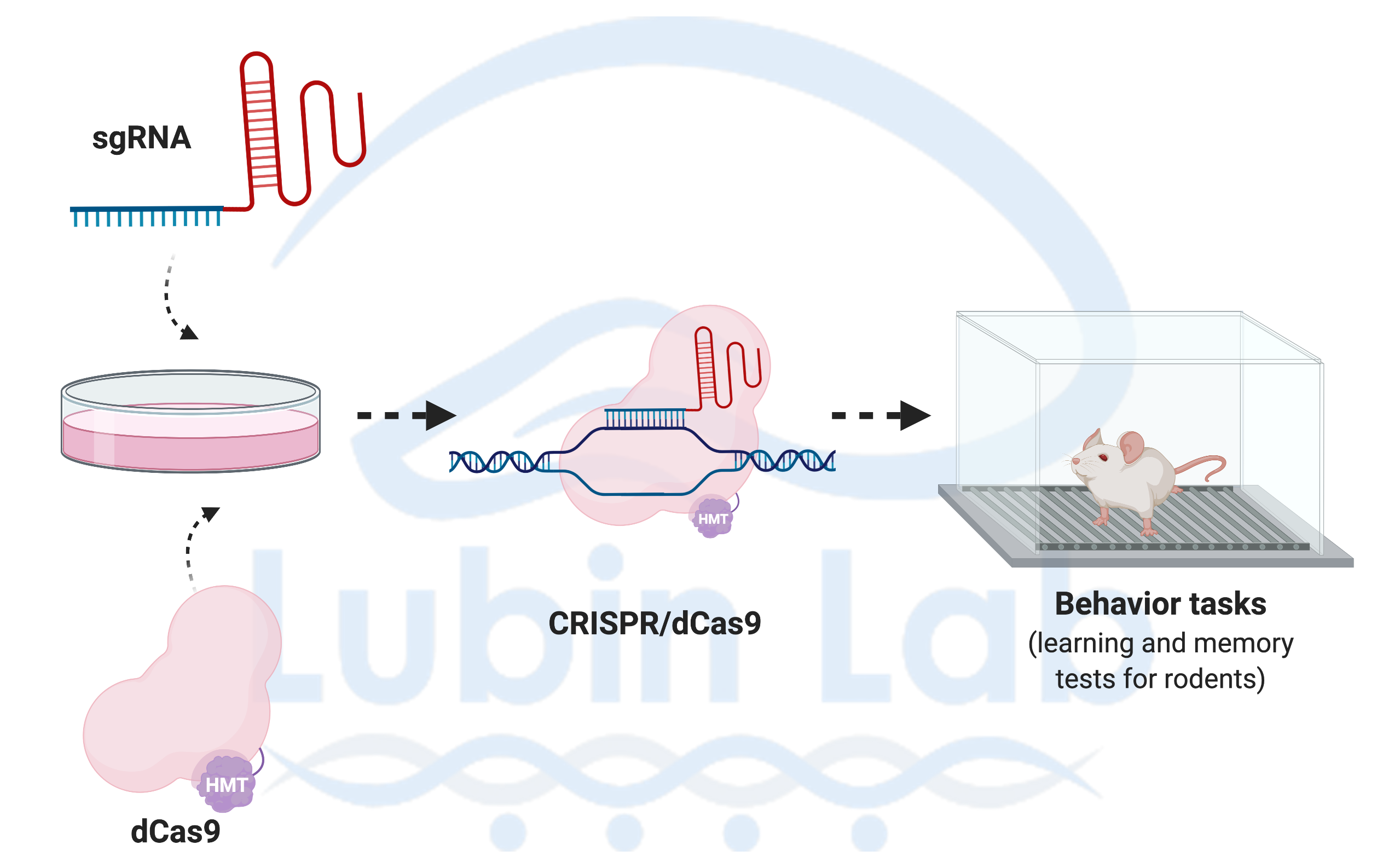
Our laboratory uses interdisciplinary methods ranging from systems to molecular neuroscience and cutting edge tools to understand how neuronal activity alters the epigenome to direct gene expression patterns: Behavior (learning and memory tasks for rats and mice); Kainate Epilepsy rodent model; Acute hippocampal slice preparations for pharmacological bath applications; Rodent brain surgeries (e.g., Cannula and electrode implantations); Gene editing [CRISPR, AAV-siRNA]; Cell and molecular biology [immunohistochemistry, quantitative real-time PCR, Methyl Specific Bisulfite PCR (MSP), Bisulfite sequencing PCR (BSP), Methyl-DNA immunoprecipitation (MeDIP), Western blotting (e.g., Histone modifications, NF-kB, p65), Electrophoretic Mobility Shift assays (EMSAs), Chromatin Immunoprecipitation (ChIP) assays, Laser-capture microdissections (LCM, e.g. cell-type specific changes in DNA methylation and gene expression (e.g. BDNF, Zif268)]; Neurophysiology (long-term potentiation); Genomic analyses (Direct Bisulfite Sequencing and Pyrosequencing); and high-throughput sequencing and bioinformatic tools to understand genome-wide transcriptional and epigenetic processes in memory disorders.
Epigenetics and Neuroplasticity - Evidence and Debate, 1st Edition is now available in print or eBook. Edited by Drs. Farah Lubin and Schahram Akbarian, the book covers topics on the entire lifespan of the brain, from transgenerational epigenetics to neurodevelopmental disease to disorders of the aging brain. All chapters are written with dual intent, to provide the reader with a timely update on the field, and a discussion of provocative or controversial findings in the field with the potential of great impact for future developments in the field. For more information go to:http://store.elsevier.com/product.jsp?isbn=9780128009772&pagename=search
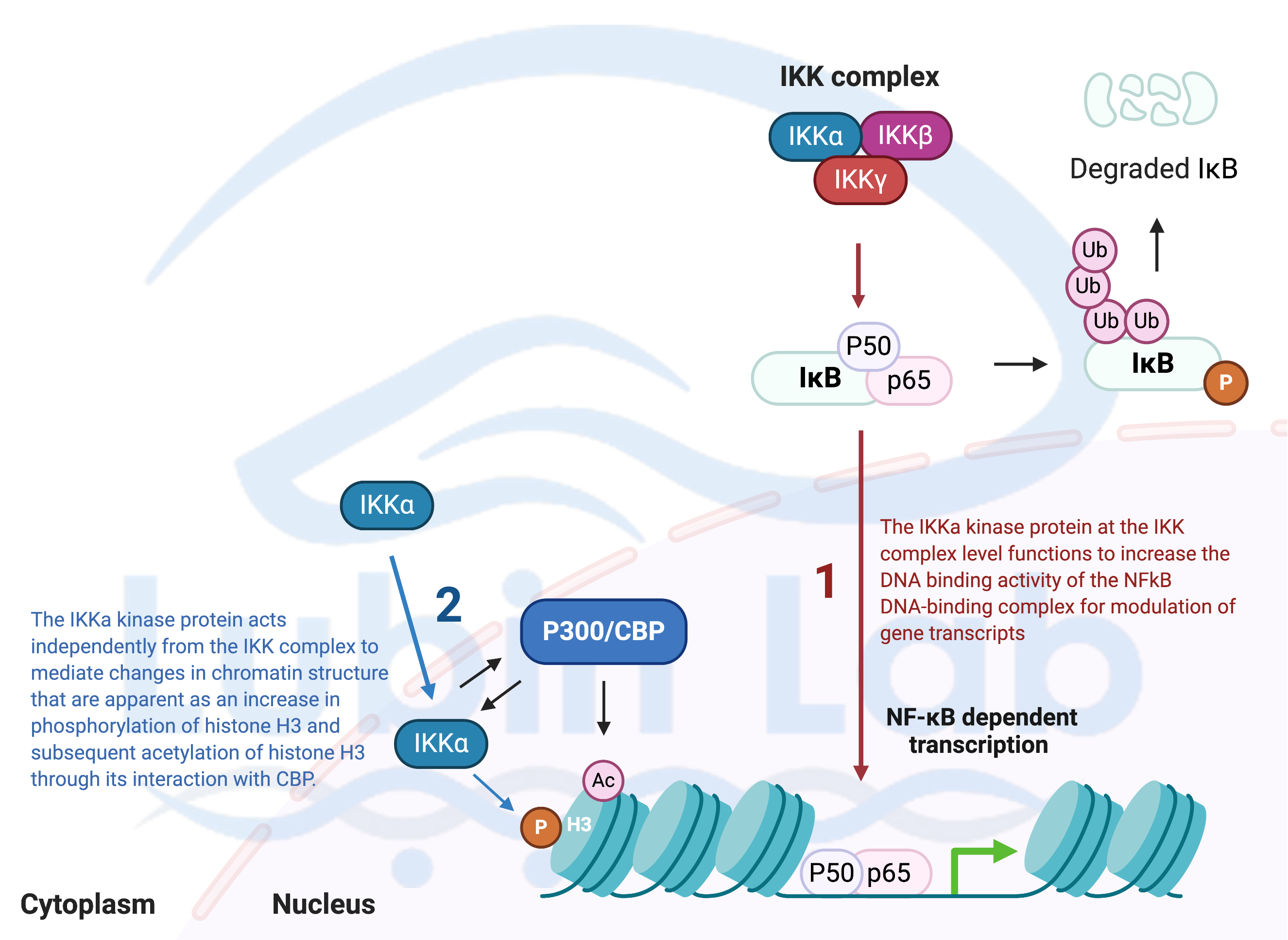
Figure 1. A Model for how Nuclear factor-kappa-light-chain-enhancer of activated B cells (NF-kB) signaling activity mediates histone H3 post-translational modification in hippocampus during memory reconsolidation (Adapted from Neuron 2007). NF-kB regulates histone phosphorylation and acetylation during memory reconsolidation.
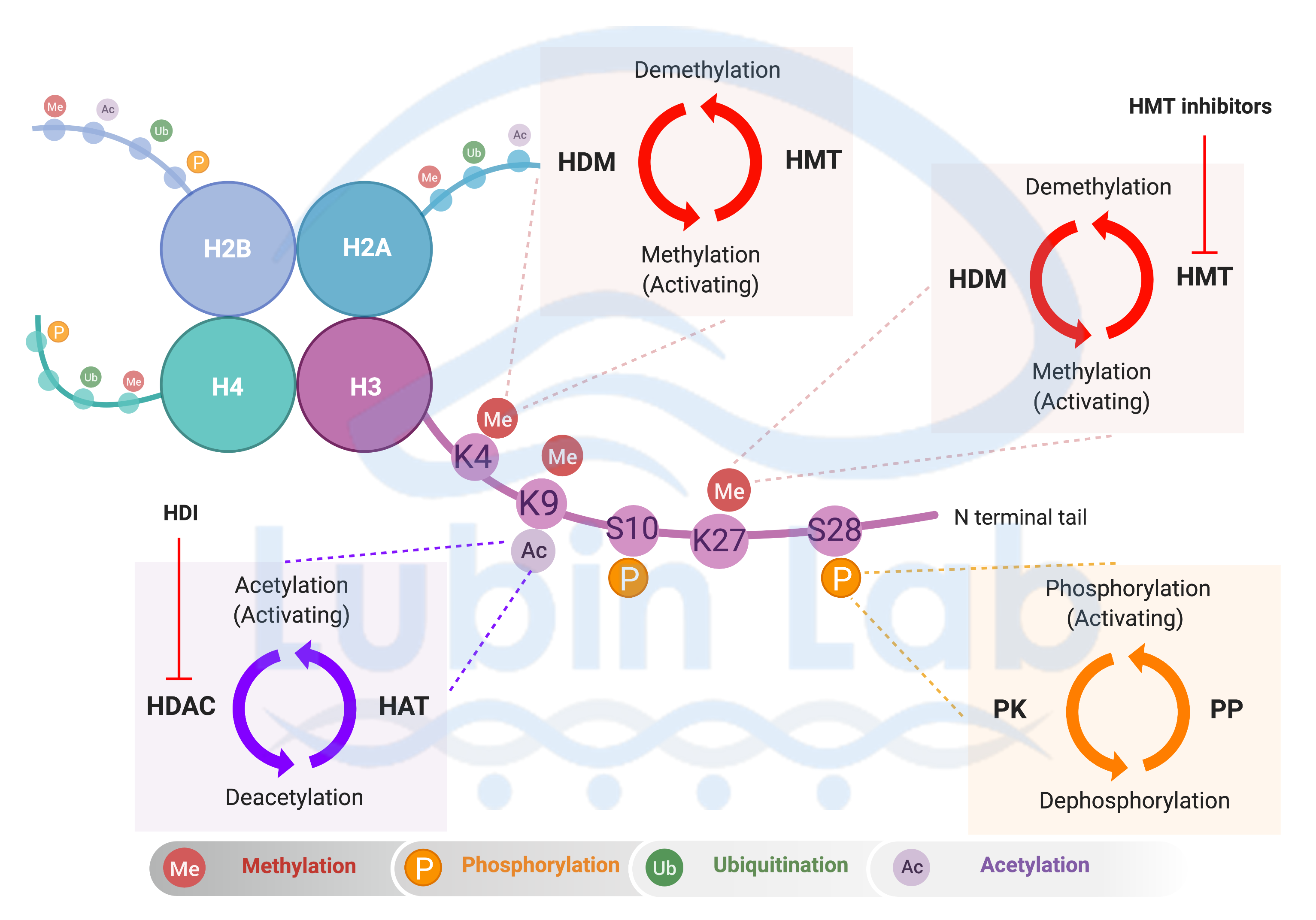
Figure 2. Model of Covalent post-translational modifications of histones in post-mitotic neurons that occur during memory formation. Schematic of the covalent histone modifications observed in histone H3, which include acetylation, phosphorylation and methylation. Lysine methylation is catalyzed by histone methyltrasferases (HMT) and the reverse reaction is catalyzed by histone demethylases (HDM). Methylation of lysine residue can result in gene activation or repression. Histone H3 Lysine acetylation is catalyzed by histone acetyl transferase (HAT) and deacetylated by histone deacetylases (HDAC). Histone deacetylase inhibitors (HDI) target HDACs resulting in de-compaction of chromatin. Serine phosphorylation is catalyzed by protein kinases (PK) and reversed by protein phosphatase (PP). H3, Histone-3; K, Lysine; S, Serine. Histone Methylation in Memory Formation
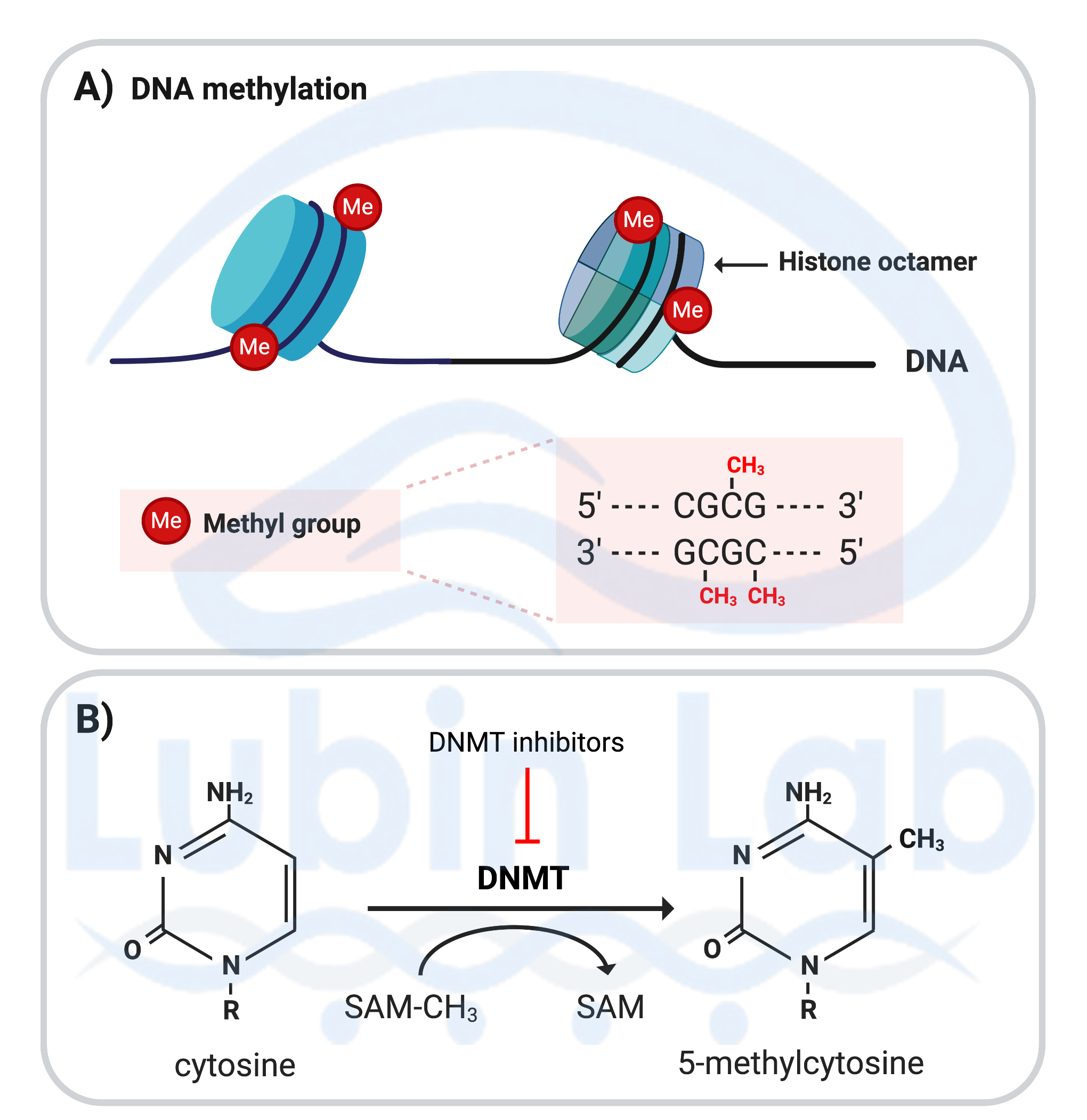
Figure 3. Covalent DNA modifications that occur in post-mitotic neurons during memory formation. A) DNA methylation occurring at cytosine residues of CpG sites, renders the DNA inaccessible to transcription. B) Mechanism of DNA methylation; the enzyme DNA methyltransferase (DNMT) catalyzes the conversion of cytosine to 5-methylcytosine. The methyl group is donated by S-adenosylmethionine (SAM). DNMT inhibitors block DNA methylation. BDNF methylation